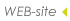|

We proposed a novel method to deliver kernels for RNA sequence data
using stochastic context free grammar (SCFG)1). Our previous
work was to deliver kernels for general biological sequences using
hidden Markov model (HMM)2). RNA sequences can not be
dealt with HMM because they involve remote base interactions which
consequently form stem-loop structures. The stem-loop structure
thermally stabilizes secondary structures of RNA, which is essential
in terms of evolutionary conservation. SCFG is more powerful stochastic
language model than HMM which allows dealing with the stem-loop
structures (Fig1). We call our novel kernel Marginalized Kernel
over SCFG. The kernel shows good performances in several demonstrations.
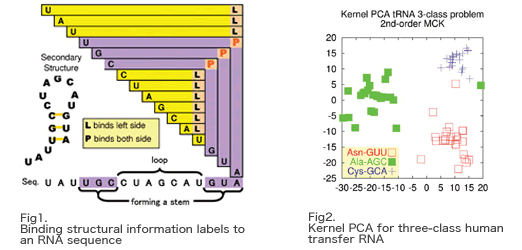
Fig2 shows a result of kernel PCA for three-class human tRNAs.
Related Information
1) T. Kin, K. Tsuda and K. Asai: "Marginalized Kernels for RNA Sequence
Data Analysis", to appear in Genome Informatics 13, 112-122 (2002)
2) K. Tsuda, T. Kin and K. Asai: "Marginalized Kernels for Biological
Sequences", Bioinformatics, Vol. 18, Suppl. 1, S268-275 (2002)

Tsuda, K., T. Kin and K. Asai: "Marginalized Kernels for Biological Sequences", Bioinformatics, Vol. 18,Suppl. 1, S268--S275(ISMB2002), 2002.
Kin, T., K. Tsuda and K. Asai,"Marginalized Kernels for RNA Sequence Data Analysis", to appear in Genome Informatics 2002.
|