|

System biology (or Systems biology) is a new frontier field of biology which investigates inside of cells as chemical reaction systems. Design principle of chemical reaction mechanism which act as life is main target of system biology.
Many reaction mechanisms are designed to describe chemical phenomena of life, such as oscillation reaction or chaos. In this research one of a general model (description) for biochemical systems is applied to gene regulatory networks in which details of molecular mechanisms are not revealed. We are developing computer algorithm for inference of network scheme of gene regulations from time series data of gene expression[1].
Generally, all chemical reaction can be described as differential equations based on mass action low or thermodynamics. For example, reaction rate will be doubled when concentration of a reactant is doubled. Although the mass action low is versatile description, it can be applicable in only case of whole reaction scheme is completely known.
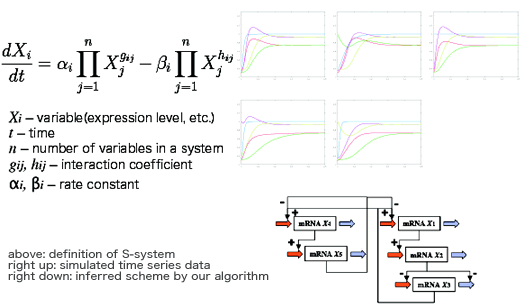
It is not suitable for biochemical systems in which reaction systems are very complex and almost unknown. For such systems, various approximations, michaeris-menten's low for enyzmic reaction, etc. are developed. The S-system[2] is one of the best representation for complex or black box systems. It is a uniform differential equation for biochemical systems including enzymic reactions. It can describe a system consist of unknown schemes, and fits for computer simulations or optimizations.
Today, while expression levels of thousands of genes are observed in time by DNA microarray technology, inference of regulatory relation among genes is very hard problem.
We are developing a algorithm for inference of gene regulatory networks only using time series data of expression levels. Our algorithm based on genetic algorithm infers network scheme from simulation data with an assumption that gene networks are scale-free network[3] like other biological networks, such as metabolic pathways or protein interaction networks.
[1] S. Kikuchi, D. Tominaga, M. Arita, K. Takahashi, M. Tomita,
"Dynamic modeling of genetic networks using genetic algorithm and
S-system", Bioinformatics, 19, 5, 643-650, 2003.
[2] M. A. Savageau, Biochemical System Analysis. A Study of Function
and Design in Molecular Biology, Addiso-Wesley, Reading, M.A., USA,
1976.
[3] J. Podani, Z.N. Oltvai, H. Jeong, B.Tombor, A.-L. Barabasi, E. Szathmary, "Comparable system-level organization of Archaea and Eukaryotes", Nature Genetics, 29, 54-56, 2001.

Kikuchi S, Tominaga D, Arita M, Takahashi K, Tomita M. "Dynamic modeling of genetic networks using genetic algorithm and S-system", Bioinformatics, 19(5), 643-650, 2003
Kadota, K., Tominaga, D., Akiyama, Y., and Takahashi, K., Detecting outlying samples in microarray data: A critical assessment of the effect of outliers on sample classification, Chem-Bio Informatics J., 3(1), 30-45, 2003.
Daisuke TOMINAGA, Katsutoshi TAKAHASHI, Nonlinear Numerical Optimization Algorithm Using System Dynamics, International Symposium on Biochemical System Theory 2002, Averoy, Norway
Daisuke TOMINAGA, "Efficient numerical optimization method for inference of interaction network structure", 2nd International Proteomics Conference (IPC2001), (poster session), (2001)
Shinichi Kikuchi, Daisuke Tominaga, Masanori Arita, Masaru Tomita, "Pathway Finding from Given Time-courses using Genetic Algorithm", the 12th International Conference on Genome Informatics(GIW 2001), (poster session), Genome Informatics Series No.12, pp.304-305, (2001)
|