|

|

|
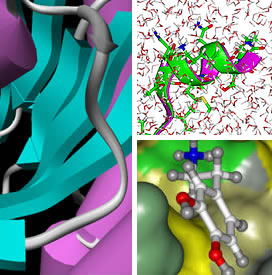
| Development
of theoretical methods for protein fold recognition and
modeling based on statistical approaches. Analysis of
the protein folding mechanisms by molecular dynamics simulation.
Ligand binding prediction and structure based drug design. |
 |
|
| Research
Interests |
|
Development of a protein fold-recognition
method and server |
|
Application to the unknown proteins
of the fold-recognition technique |
|
Application to the unknown proteins
of the fold-recognition technique |
|
Protein folding analysis by
Multicanonical Molecular Dynamics
1:Energy landscape of a peptide
cut from a distal beta-hairpin
of src SH3 domain
2:Energy landscape of a "Chameleon"
sequence and its bilateral property |
|
Analysis of a reentrant loop
in membrane proteins for classification
and prediction |
|
Comparative Modeling of Class
A Rhodopsin like GPCRs |
|
Comparative ligand-binding analysis
for molecular modeling of the
binding of ligands to a receptor |
|
|
|
|
|
 |
|
 |
|
Team Leader : Takatsugu
Hirokawa
|
|
|
| |
|
| Name
|
Keywords |
Reference
|
|
Tomoshi Kameda
AIST Research Staff
|
molecular dynamics, replica exchange method, protein folding, amyloid and aggregation |
|
|
Nobuyuki Uchikoga
AIST Research Staff
|
|
|
| |
|
- Joint research
- Tsumura & Co.
- Mochida Pharmaceutical Co., Ltd.
- Taiho Pharmaceutical Co., Ltd.
- Technical Training
- Ajinomoto Co., Inc.
- Ryoka Systems Inc.
- Mizuho Information & Research Institute, Inc.
- Mitsui Knowledge Industry Co., Ltd.
|
|